complete = read.csv(yourfiles[1])
for(i in 2:length(yourfiles)){
part = read.csv(yourfiles[i])
complete = merge(complete, part, all=TRUE, sort=FALSE)
}
It wasn't really a problem back in the summer because I didn't have to think it that much. But now it's realy a pain as I need to merge over 160 000 lines. So because I needed to save some time I came up with the following merging algorithm:
#data.list = list of your frames you want to merge
#nparts = how many parts are merged inside apply, the optimal value is probably
#different for every machine, but should not be a big deal.
fast.merging = function(data.list, nparts=10){
if(!is.list(data.list)) stop("data.list isn't a list") #standard error
while(length(data.list) != 1){ #Loop until everything is merged
#define sections according to nparts
if(length(data.list) > nparts){
starts = seq(1, length(data.list), nparts)
ends = seq(nparts, length(data.list), nparts)
#starts and ends are of equal size
#if length(data.list) divides nparts.
if(length(ends) < length(starts)) ends = c(ends, length(data.list))
#making sure things are even
sections = matrix(c(starts, ends), ncol=2, byrow=FALSE)
sections = apply(sections, 1, list)
}else{
sections = list(c(1, length(data.list)))
}
#We have the standard way inside lapply
#lapply merges a length(sections) amount of nparts sections
data.list = lapply(sections, function(x, data.list){
if(is.list(x)) x = x[[1]]
#the standard way starts ->
part = data.list[[x[1]]]
for(i in x[1]:x[2]){
part = merge(part, data.list[[i]], all=TRUE, sort=FALSE)
}
#<- standard way ends
return(part)
}, data.list = data.list)
#rinse and repeat until all is done
}
return(data.list[[1]]) #returning the merged data frame
}
The scheme seems completely unintuitive at first. Because you are basically using lapply to vectorize for-loop merging. But it probably makes sense when you have a grasp how vectorizing works in R, or why you should always define the size of your variables before you use them. But enough of that, time for the actual benchmark results:
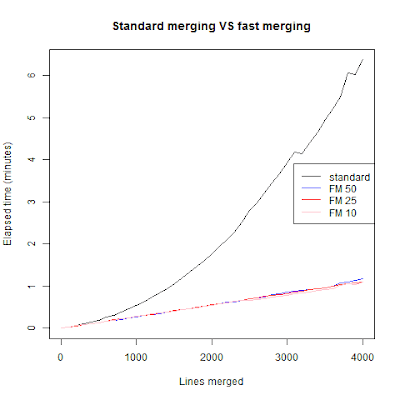
As we can see the standard for-loop merging grows exponentially in time as we merge more frames. The algorithm I used grows in linear time and didn't really depend on the nparts argument. Meaning that with nparts being 50, 25 or 10 the time difference is few seconds when we are at 4001 merges. I didn't want to go further because that for-looping get's really tedious to wait. I think we can do better then fast merging, for one we can use parallel calculations. But that's for next week (read: hopefully this weekend).
Benchmark was created with this scheme:
#Getting our lines to a list of lines
#my frames, in this case only line per data-frame.
eve.frames = lapply(eve.list, eve.data.frame) #Some Eve Online kill mails
sekvenssi = seq(1, 4001, 100) #only trying 1, 101, 201, ... 4001.
time1 = rep(NA, length(sekvenssi))
for(i in 1:length(sekvenssi)){
complete = eve.frames[[1]]
time1[i] = system.time(
for(j in 2:sekvenssi[i]){
complete = merge(complete, eve.frames[[j]],
all=TRUE, sort=FALSE)
}
)[3]
print(paste(round(time1[i],2), nrow(complete),i))
}
write(time1, "time1.dat")
#backup = complete
time2 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time2[i] = system.time(fast.merging(complete, 50))[3]
print(paste(round(time2[i],2), nrow(complete),i))
}
write(time2, "time2.dat")
time3 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time3[i] = system.time(fast.merging(complete, 25))[3]
print(paste(round(time3[i],2), nrow(complete),i))
}
write(time3, "time3.dat")
time4 = rep(NA, length(sekvenssi))
for(i in seq(sekvenssi)){
complete = eve.frames[1:sekvenssi[i]]
time4[i] = system.time(fast.merging(complete, 10))[3]
print(paste(round(time4[i],2), nrow(complete),i))
}
write(time4, "time4.dat")
#To check if they are correct try
#complete = fast.merging(complete, 10)
#backup = backup[order(backup[, 1]), order(colnames(backup))]
#complete = complete[order(complete[, 1]), order(colnames(complete))]
#sum(backup == complete, na.rm=TRUE)
#sum(is.na(backup))
#these should add up to the matrix size.
plot(sekvenssi, time1/60, type="l", ylab="Elapsed time (minutes)", xlab="Lines merged", main="Standard merging VS fast merging")
points(sekvenssi, time2/60, type="l", col="blue")
points(sekvenssi, time3/60, type="l", col="red")
points(sekvenssi, time4/60, type="l", col="pink")
legend("right", c("standard", "FM 50", "FM 25", "FM 10"),
lty=c(1,1,1,1), col=c("black", "blue", "red", "pink"))
Very interesting benchmark.
ReplyDeleteWould you mind to give the data.table package a try and benchmark also the data.table merge() ?
Brgds,
blu
Definetely, I will do it this weekend!
Delete- Xachriel
This really helps. Much faster even using a list of data tables. I added another parameter for the keys to use when merging and the chunk approach reduced the time from over an hour to 2 minutes. Thanks.
ReplyDelete